KRAS-related Assays: Binding kinetics of agents to K-Ras are measured via surface plasmon resonance (SPR) technique
K-Ras
KRAS
K-Ras fragments aa 2-169 with a C-terminal biotinylation expressed in E. coli. Data shown here was collected for KRAS in a GDP-bound state.
C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, KRAS1, KRAS2, NS, NS3, RASK2
K-Ras: P01116 (https://www.uniprot.org/uniprot/P01116)
Small molecule and peptide binding to KRAS wildtype and mutants.
Malvern, PA, USA
More information can be found on our website SPR Assay Services for Drug Discovery.
Binding kinetics of KRpep-2d to wild type K-Ras and mutants G12C, G12D, G12V, and G12R
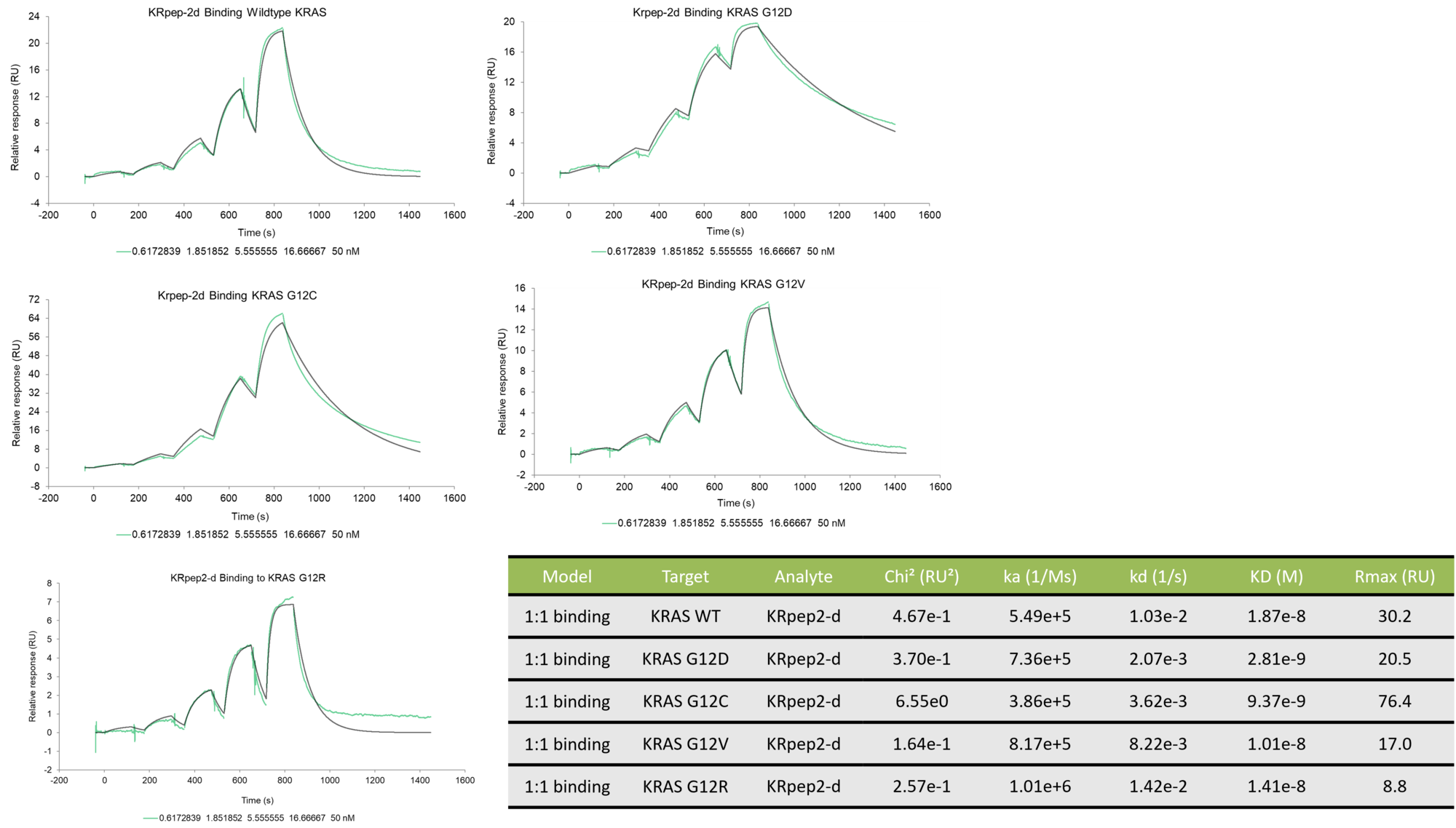
The data was collected on a Biacore 8K using single-cycle kinetics. The data was fit using a 1:1 kinetic binding model. The peptide binds with the tightest affinity to K-Ras mutant G12D.
Binding kinetics of BI-2852 to wild type K-Ras and mutants G12C, G12D, G12V, G12R, and double mutant G12D/T35S
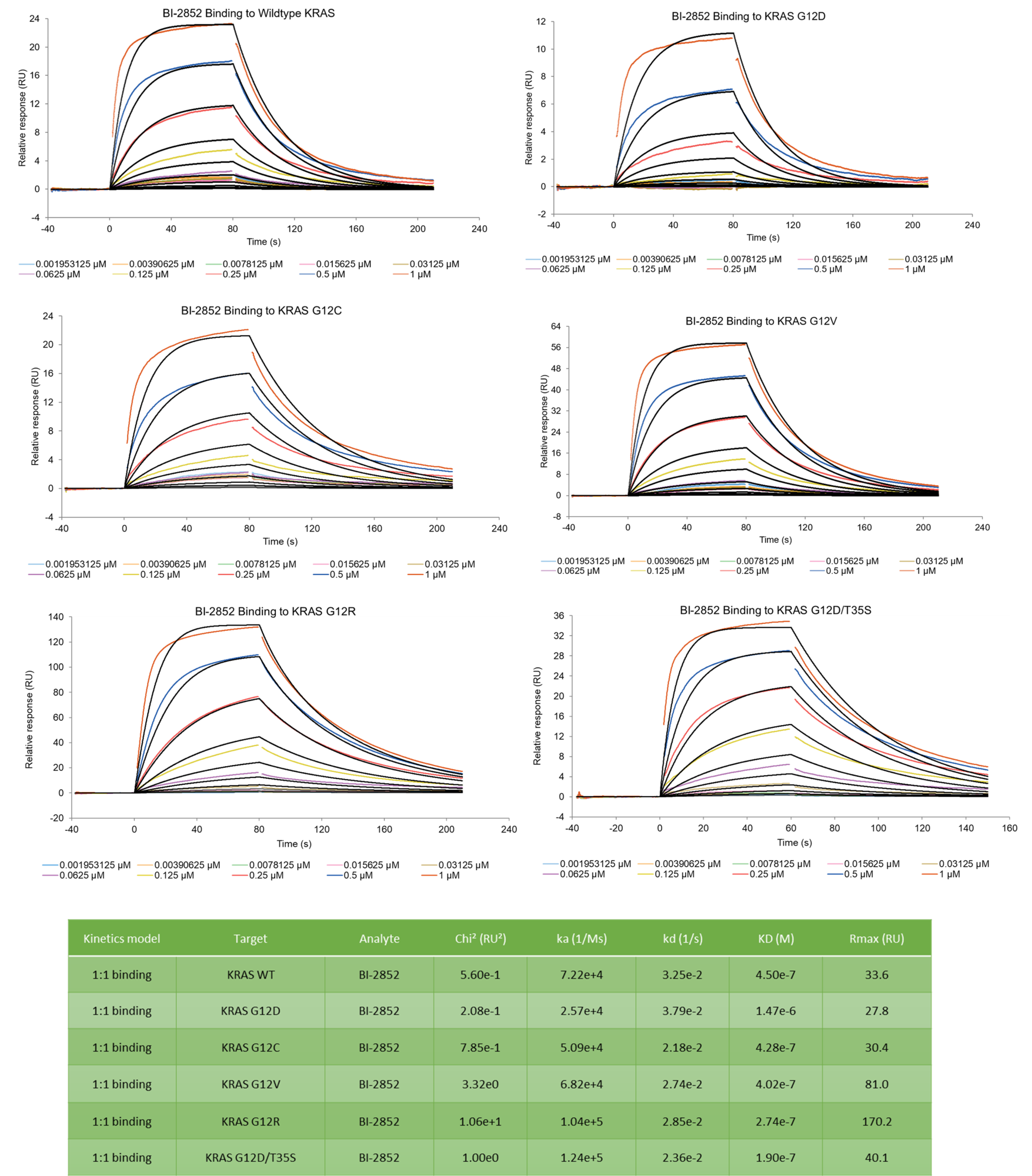
The data was collected on a Biacore 8K using multi-cycle kinetics. The data was fit using a 1:1 kinetic binding model. BI-2852 binds to all KRAS constructs with similar kinetics/affinity.
Binding kinetics of MRTX1133 to wild type K-Ras and mutants G12C, G12D, and G12V
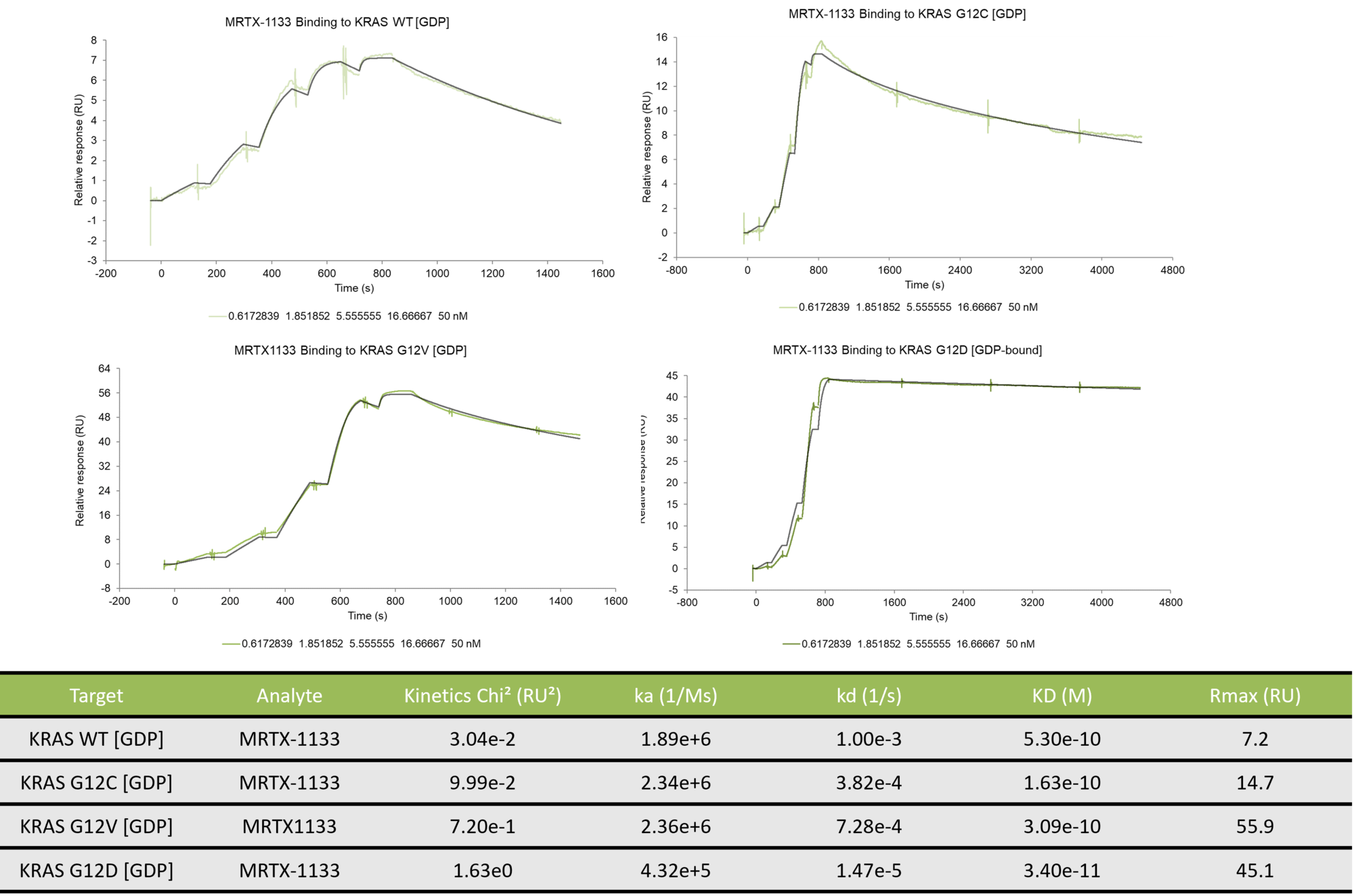
The data was collected on a Biacore 8K using single-cycle kinetics. The data was fit using a 1:1 kinetic binding model. While MRTX1133 exhibits relatively tight binding to all the KRAS constructs tested, the off-rate is significantly slower for binding to KRAS G12D resulting in an KD value that is at least an order of magnitude tighter than for the other constructs.