Spotlight: Cell-based kinase assay formats.
Running cell-based assays in multiple ways provides an opportunity to view a kinase inhibitor in action from different angles. At Reaction Biology, we offer three assay formats for cell-based kinase screening. In this blog, we compare the assay formats to help you choose the assay that is right for your research project.
Reaction’s Assay Formats
The NanoBRET Intracellular Target Engagement Assay technology enables the investigation of compound-target engagement (or binding) in the physiological environment of intact cells. NanoBRET is an energy transfer technique designed to measure the apparent affinity of test compounds by competitive displacement of a NanoBRET tracer, reversibly bound to a NanoLuc luciferase-kinase fusion construct expressed in cells. Upon binding of the drug to the target kinase, the fluorescent tracer is pushed away from the binding site, increasing the tracer’s proximity to the luciferase-kinase construct, resulting in a decreased BRET signal.
The Cellular Phosphorylation Assay is an assay for quantification of the activity of a kinase. The assay is performed with cells either expressing the endogenous kinase or a kinase construct. Substrate phosphorylation is quantified via detection with phospho-site specific antibodies. Upon kinase inhibition by a compound, the kinase stops phosphorylating the substrate, which is quantified as a readout for kinase activity.
The BaF3 Cell Proliferation Assay, which is often called BaF3 Cell Transformation Assay, is an activity assay format designed to examine the transformation capacity of a kinase. The survival of BaF3 cells is dependent on signaling from extracellular Interleukin-3 (IL3). This dependency can be overcome by the expression of a transforming kinase — the expression of an oncogenic kinase that drives cell proliferation even in the absence of IL3. Inhibition of the kinase by a compound deprives the cells of the oncogenic driver resulting in cell death.
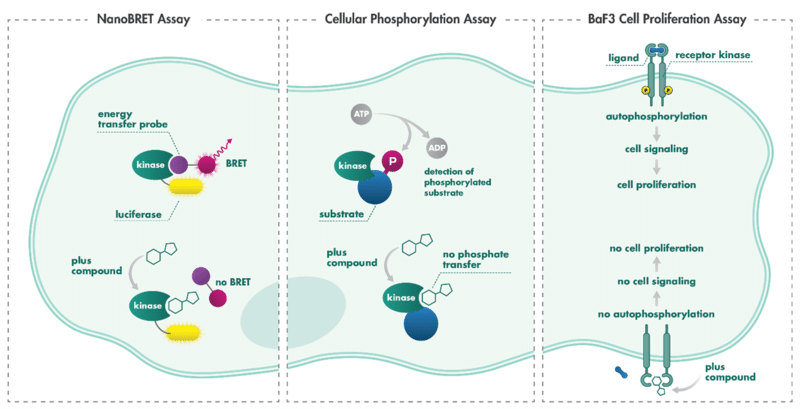
Comparing the Cellular Phosphorylation Assay and NanoBRET Intracellular Target Engagement Assay
It is recommended to conduct enzyme activity assays that quantitate substrate phosphorylation as well as target engagement assays that measure binding affinity and target residence time. Neither approach is flawless, but when used together, potential methodological shortcomings are easier to identify. One limitation of the NanoBRET assay is that since all the constructs are exogenous, the abundance of the target may not be physiological. In addition, while it has been shown that inhibitors of all classes, including allosteric inhibitors, can be detected using NanoBRET technology, the assay outcomes could be influenced by the tracer’s binding mode (Vasta et al.1).
On the other hand, the Cellular Phosphorylation Assay reports percent of inhibition of kinase activation and IC50. These assays use an ELISA or AlphaLISA method that utilizes target-specific antibodies and phospho-site-specific antibodies. The sandwich principle is used to further increase selectivity by requiring the detection of two independent sites of the same protein. However, there will always be some time lag between kinase inhibitor-mediated inhibition and the subsequent change in the detectable phosphorylation status of the chosen substrate. This is due to the preexisting baseline level of endogenous substrate phosphorylation, as well as intracellular processes including substrate turnover, phosphatase activities, or autophosphorylation (Ouellette et al.2). Kinases may have multiple substrates and some kinases’ native substrates are still not identified, therefore, developing cellular phosphorylation assays against new kinase targets can be more challenging. In contrast, performing target engagement assay with an overexpressed kinase will provide a relatively quick answer.
Ideally, investigations of binding interactions and physiological outcomes should both be undertaken within a cellular context. Towards that end, the NanoBRET platform assesses target engagement in intact cells and provides real-time data of the binding affinity as well as target residence time.
Drug-target residence time is the reciprocal of the dissociation rate constant of a bound ligand and reflects the lifetime of the drug-target complex. While sometimes correlated with affinity (Kd or IC50), a decrease in the dissociation rate constant does not always go hand-in-hand with increased potency proxies.
For example, while the inhibitor lapatinib binds about 10-fold less potently to EGFR than gefitinib (Ki app values of 3 and 0.4 nM, respectively), the residence time of lapatinib on EGFR is 30-fold longer. This increased residence time for lapatinib was associated with prolonged inhibitory activity in cells. The reason is that although the thermodynamic selectivity is identical, if the association constant and dissociation constant are different, then there will be some kinetic selectivity. Similarly, if the IC50 of a particular compound is the same for both the target and some off-target protein, it is still possible for the compound to be selective.
The kinetic selectivity will come into play when talking about compounds that eliminate with different half-lives. For example, a drug with a half-life of 5 h may reach >95% occupancy at a low initial drug dose at all its potential targets, while a different drug with similar thermodynamic selectivity but a half-life of 1 h might still be at just 5% occupancy for one target while saturating the others (Tonge et al.3).
When should you use the Cellular Phosphorylation Assay vs. BaF3 Cell Proliferation Assay?
The Cellular Phosphorylation Assay specifically focusses on the impact of kinase inhibition on the phosphorylation of exactly one downstream substrate which is affected within a short time frame of 90 min. The assay can be performed in the absence of serum, excluding the impact of serum-binding to the interpretation of the inhibitor activity.
The BaF3 system relies on the principle that normal BaF3 cells die after the withdrawal of IL-3. Transduction of the cells with an oncogenic kinase construct establishes BaF3 cell proliferation independent of IL-3, now reliant on the kinase as oncogenic driver.
Cell death of kinase driver-addicted BaF3 cells due to kinase inhibition can be rescued by supplementing IL-3, thus providing a counter screen for off-target effects. Such drug studies can determine the potency of compounds against wild type or mutant kinases as well as inhibition resistant alleles, predict and confirm compound-target binding modes, and reveal new therapeutic liabilities for oncogenic variants (Kong et al.4). More uses of the BaF3 Cell Proliferation Assay for kinase drug discovery are outlined in a review by Warmuth et al.5.
The BaF3 assay uses cell proliferation over several days as the readout, requires serum presence, and analyses the impact of compounds on the activated pathway in general.
The BaF3 system is helpful to:
- Access the transforming capacity of new kinase mutations or kinase fusion proteins
- Predict resistance to small molecule kinase inhibitors elicited by point mutations
- Determine kinase inhibitor potency
- Investigate the signaling events downstream of the kinase of interest
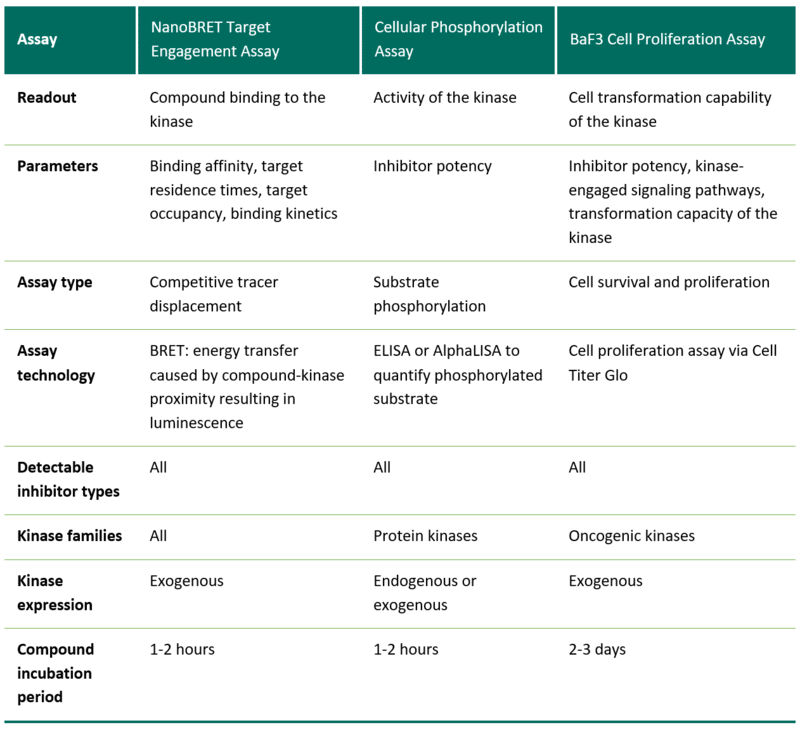
Which kinases can be investigated with the respective assay formats?
The NanoBRET Intracellular Target Engagement Assay is performed using kits from Promega. Assays for more than 350 kinases are currently available (as of March 2022).
The Cellular Phosphorylation Assay can be established when a substrate is known to be regulated by the kinase of interest and the substrate is expressed at sufficient levels for detection. To ensure its high specificity, the detection requires the existence of two substrate-specific antibodies, one of which is phospho-site-specific.
The BaF3 Cell Proliferation Assay can be performed with kinases that have the ability to drive cell proliferation after stable cell transduction, i.e., oncogenic kinases.
Read more about the importance of screening compounds in cell-based assays vs. biochemical assays in our blog: Testing kinase inhibitors where it matters: Drug screening in intact cells.
